PhAST: Physics-Aware, Scalable, and Task-specific GNNs for Accelerated Catalyst Design#
This repository contains implementations for 2 of the PhAST components presented in the paper:
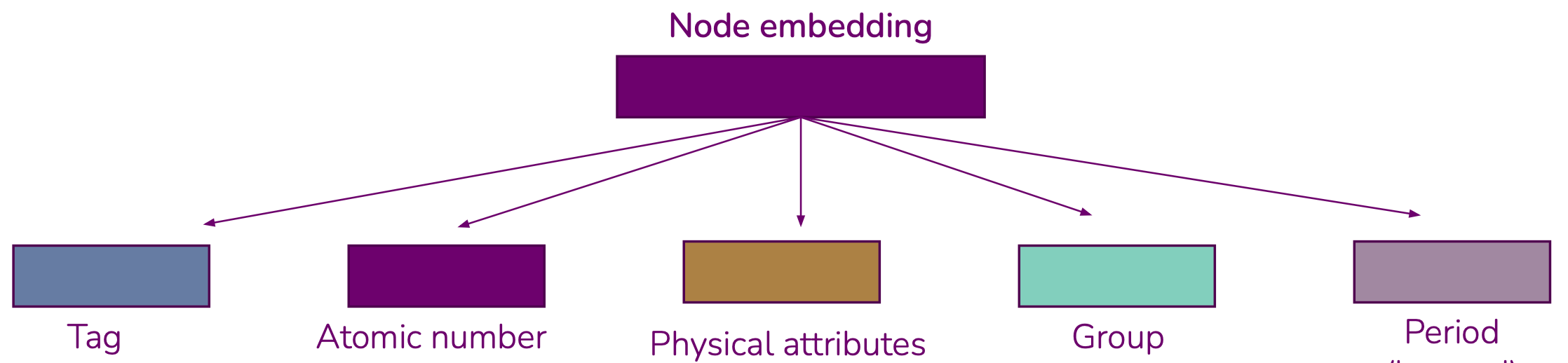
PhysEmbeddingthat allows one to create an embedding vector from atomic numbers that is the concatenation of:A learned embedding for the atom’s group
A learned embedding for the atom’s period
A fixed or learned embedding from a set of known physical properties, as reported by
mendeleevIn the case of the OC20 dataset, a learned embedding for the atom’s tag (adsorbate, catalyst surface or catalyst sub-surface)
Tag-based graph rewiring strategies for the OC20 dataset:
remove_tag0_nodesdeletes all nodes in the graph associated with a tag 0 and recomputes edgesone_supernode_per_graphreplaces all tag 0 atoms with a single new atomone_supernode_per_atom_typereplaces all tag 0 atoms of a given element with its own super node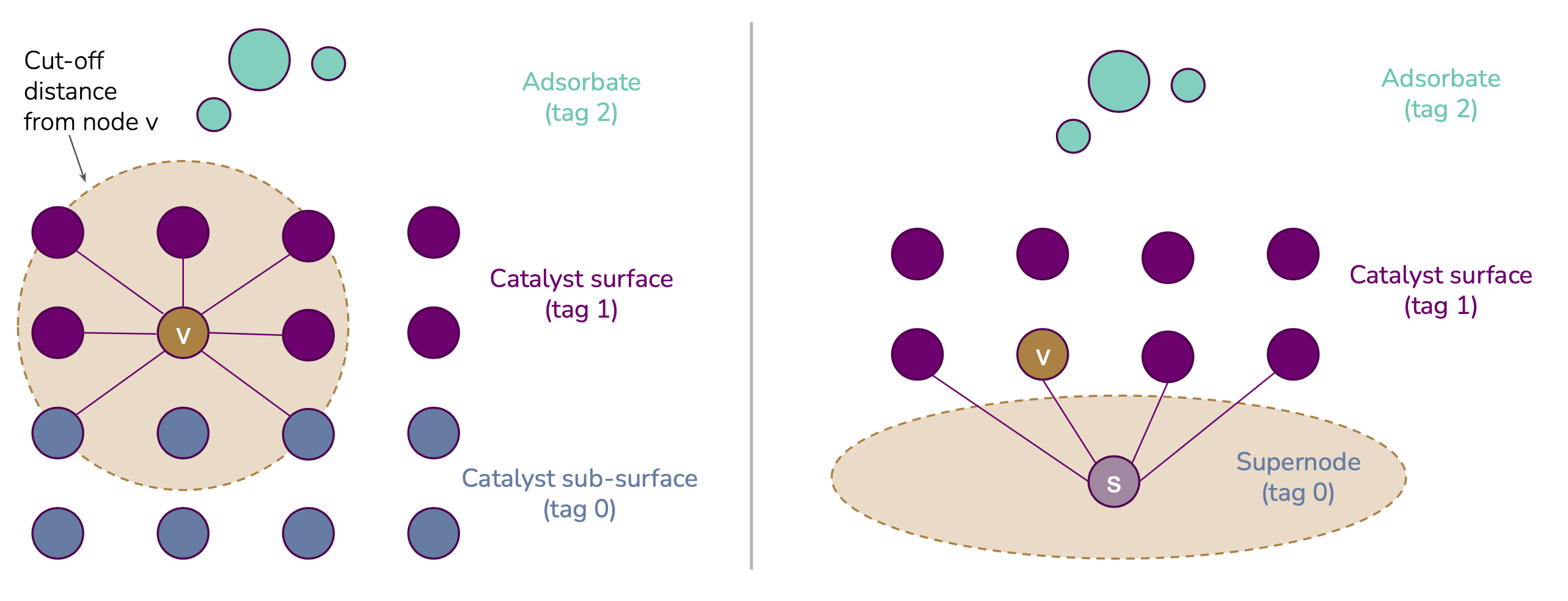
Also: https://github.com/vict0rsch/faenet
Installation#
pip install phast
⚠️ The above installation does not include torch_geometric which is a complex and very variable dependency you have to install yourself if you want to use the graph re-wiring functions of phast.
☮️ Ignore torch_geometric if you only care about the PhysEmbeddings.
Getting started#
Physical embeddings#
import torch
from phast.embedding import PhysEmbedding
z = torch.randint(1, 85, (3, 12)) # batch of 3 graphs with 12 atoms each
phys_embedding = PhysEmbedding(
z_emb_size=32, # default
period_emb_size=32, # default
group_emb_size=32, # default
properties_proj_size=32, # default is 0 -> no learned projection
n_elements=85, # default
)
h = phys_embedding(z) # h.shape = (3, 12, 128)
tags = torch.randint(0, 3, (3, 12))
phys_embedding = PhysEmbedding(
tag_emb_size=32, # default is 0, this is OC20-specific
final_proj_size=64, # default is 0, no projection, just the concat. of embeds.
)
h = phys_embedding(z, tags) # h.shape = (3, 12, 64)
# Assuming torch_geometric is installed:
data = torch.load("examples/data/is2re_bs3.pt")
h = phys_embedding(data.atomic_numbers.long(), data.tags) # h.shape = (261, 64)
Graph rewiring#

from copy import deepcopy
import torch
from phast.graph_rewiring import (
remove_tag0_nodes,
one_supernode_per_graph,
one_supernode_per_atom_type,
)
data = torch.load("./examples/data/is2re_bs3.pt") # 3 batched OC20 IS2RE data samples
print(
"Data initially contains {} graphs, a total of {} atoms and {} edges".format(
len(data.natoms), data.ptr[-1], len(data.cell_offsets)
)
)
rewired_data = remove_tag0_nodes(deepcopy(data))
print(
"Data without tag-0 nodes contains {} graphs, a total of {} atoms and {} edges".format(
len(rewired_data.natoms), rewired_data.ptr[-1], len(rewired_data.cell_offsets)
)
)
rewired_data = one_supernode_per_graph(deepcopy(data))
print(
"Data with one super node per graph contains a total of {} atoms and {} edges".format(
rewired_data.ptr[-1], len(rewired_data.cell_offsets)
)
)
rewired_data = one_supernode_per_atom_type(deepcopy(data))
print(
"Data with one super node per atom type contains a total of {} atoms and {} edges".format(
rewired_data.ptr[-1], len(rewired_data.cell_offsets)
)
)
Data initially contains 3 graphs, a total of 261 atoms and 11596 edges
Data without tag-0 nodes contains 3 graphs, a total of 64 atoms and 1236 edges
Data with one super node per graph contains a total of 67 atoms and 1311 edges
Data with one super node per atom type contains a total of 71 atoms and 1421 edges
Tests#
This requires poetry. Make sure to have torch and torch_geometric installed in your environment before you can run the tests. Unfortunately because of CUDA/torch compatibilities, neither torch nor torch_geometric are part of the explicit dependencies and must be installed independently.
git clone git@github.com:vict0rsch/phast.git
poetry install --with dev
pytest --cov=phast --cov-report term-missing
Testing on Macs you may encounter a Library Not Loaded Error
Requires Python <3.12 because
mendeleev (0.14.0) requires Python >=3.8.1,<3.12


